AI Breakthrough Unmasks Hidden Threat of Superbugs | Image Source: medicalxpress.com
NEW ORLEANS, Louisiana, April 2, 2025 – As drug-resistant infections continue to intensify in what many see as a “silent pandemic,” scientists and health professionals around the world are mobilizing powerful new tools to convert the tide. Among the most promising advances is a pioneering IA method at Tulane University that can revolutionize how physicians detect antibiotic resistance in bacteria, a critical factor in treatment success or failure. Supported by comprehensive international research and validated with clinical samples, the model’s performance exceeds current gold standards and offers the hope of faster and more accurate responses to one of the most serious threats in medicine.
According to the World Health Organization (WHO), drug-resistant infections are among the main threats to global public health, with MDR TB affecting 450,000 people in 2021. In many cases, survival is a coin launch: treatment success rates decreased to only 57% that year. The crisis is the excessive use of antibiotics in medicine and agriculture, which accelerates the evolution of resistant strains. These superchips make conventional antibiotics useless, prolong diseases, increase health costs and increase mortality rates. But if the Tulane University Group (GAM) model remains a success, chances can finally begin to build on humanity.
What is the new GAM+ machine learning approach?
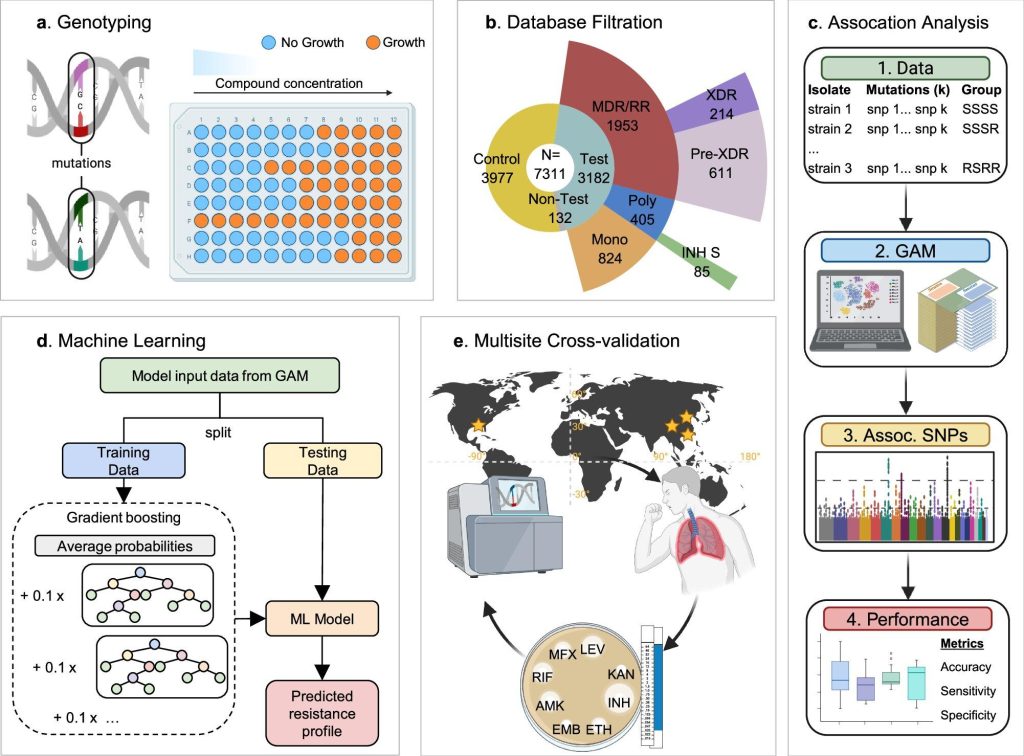
The new technique introduced by researchers Tulane is married to a group association model (GAM) with automatic learning to discover subtle yet significant genetic models that indicate drug resistance in bacteria. Unlike traditional tools that depend on predefined knowledge of genes causing resistance, this method uses data-based algorithms to make discoveries that even experienced microbiologists could lose. As indicated by Nature Communications, where the research was published, the model analyses the entire genome of bacterial strains – including Mycobacterium tuberculosis (Mtb) and Staphylococcus aureus – and compares resistance profiles to detect true culprits behind antibiotic failure.
“We essentially teach a computer to recognize resistance models without having to report them first,” said Dr. Tony Hu, the author and principal director of the Tulane Center for Cell and Molecular Diagnostics. The model was tested in more than 7,000 TB strains and about 4,000 staf samples, where the accuracy of the current WHO genotypic resistance detection databases coincided or exceeded, while reducing the risk of false positives, a problem that can lead to poor prescribed drugs, often with fatal consequences.
Why is accurate detection so important?
The main problem with the current resistance detection is in two bottlenecks: time and accuracy. Culture-based testing may take weeks to produce results, while DNA-based techniques may lose rare or rare mutations. Meanwhile, doctors are forced to rely on antibiotics or wide-spectrum puzzles, which can increase resistance over time. According to Julian Saliba, principal author and graduate student in Tulane, the GAM model considerably reduces diagnostic noise. “Our method provides a clearer picture of which mutations really cause resistance, reducing misdiagnosis and unnecessary changes to treatment”
he said.
Clinical validations in China have already shown that the GAM + ML model exceeds WHO methods for predicting resistance to first-line antibiotics. This type of precision means that treatment plans can be adapted more effectively from the outset, which could reduce hospital stays and improve patient outcomes, while reducing the spread of antibiotic resistance.
Can I help you beyond hospitals?
Interestingly, the potential of this tool does not stop at the clinic door. According to the research, its adaptability could make it useful in sectors such as agriculture, where the abuse of antibiotics in livestock contributes greatly to the crisis of resistance. In the analysis of bacterial strains in agricultural environments, AI tools such as GAM could help limit exposure to antibiotics in food chains, a problem that has already caused regulatory interventions in several countries. It could also be deployed in monitoring wastewater treatment plants, one of the main points of contact for gene transfer between bacteria, according to researchers at the University of Technology in Chalmers, Sweden.
“We find that bacteria found in water and human treatment plants are more likely to become resistant by gene transfer,” said Erik Kristianson, Professor of Chalmers, whose independent research also applied AI to track how resistance genes jump among bacterial species. His model, composed of a data set composed of nearly one million bacterial genomes, confirmed that genetic similarity increases the possibilities for transfer of resistance genes, a discovery that could redefine how and where to monitor by emerging threats.
What role does New Zealand play?
On the other side of the world, New Zealand is taking a proactive attitude with its own digital approach to antimicrobial resistance. As part of a $36 million project funded by the Te Niwha government, researchers are building the first national digital antimicrobial guidelines platform. According to Dr. Stephen Ritchie, Associate Professor at Auckland University and Chair of the Antimicrobial Stewardship Committee at Auckland Hospital, aims to unify fragmented practices of antibiotic prescriptions among all health care providers.
“These guidelines vary in quality and remain up to date, and what is equivalent to variable treatments,” said Ritchie. He stressed the urgent need for coherence, explaining that unified numerical guidelines could reduce unnecessary or ineffective requirements, a central engine of resistance. Tim Packer, the emerging technology director of RUSH Digital, the technology company involved in the project, stressed that their previous experience in the development of the Covid Tracer app has placed them well in order to offer once again impressive health technology solutions.
How does resistance spread so quickly?
Resistance is not random – it thrives in environments where bacteria are forced to evolve. Hospitals, farms and sewer systems are zero-earth. In these adjustments, antibiotics are often used, with a selection pressure that eliminates sensitive bacteria and promotes resistant bacteria. Add to this the fact that bacteria can exchange resistance genes like business cards through a process called horizontal gene transfer, and you have a recipe for superbsugs. AI models such as those developed by Tulane and Chalmers help decipher this complex process, providing a clearer view of when, where and how these genes jump between organisms.
“Most resistance genes are shared between bacteria with similar genetic structures,” says Kristianson. “We think this reduces the cost of taking new genes.” When resistance becomes “affordable” in the evolutionary sense, it extends more effectively. The Chalmers model, consisting of huge data sets and tested by blind validation, accurately transfers the resistance gene predicted four times five times. Future iterations, formed in even larger data sets, could be even more accurate.
Can AI become a front-line diagnostic tool?
The vision of IV in diagnosis is ambitious and necessary. As David Lund, a medical researcher at Chalmers, said, the ultimate goal is to integrate these models into real-time diagnostic platforms, systems that could, for example, analyze a patient’s sample and instantly determine if a superbug is present and resistant to standard treatments. This would not only help physicians make immediate decisions, but could also serve public health organizations by marking new points of resistance before epidemics become out of control.
“Now we can really work with data to answer complex questions,” said Kristianson. But more importantly, we can ask new questions. This paradigm shift, driven by AI and mass data sets, could define the next decade of infectious disease control, especially when traditional antibiotics lose their effectiveness. Since the streamlining of hospital workflows to guide environmental monitoring and to report on global health policy, the future of resistance management can be modelled less by the laboratory bank and even more by the algorithm.
Although the bets are high and the way forward remains uncertain, one thing is clear: doing nothing is not an option. Whether it is a model for digitizing millions of genomes or a national platform that harmonizes the guidelines, each new tool brings us closer to the changes that threaten to make modern medicine ineffective. As infections evolve, they also need our tools. And for now, at least it seems that AI is helping us evolve faster.